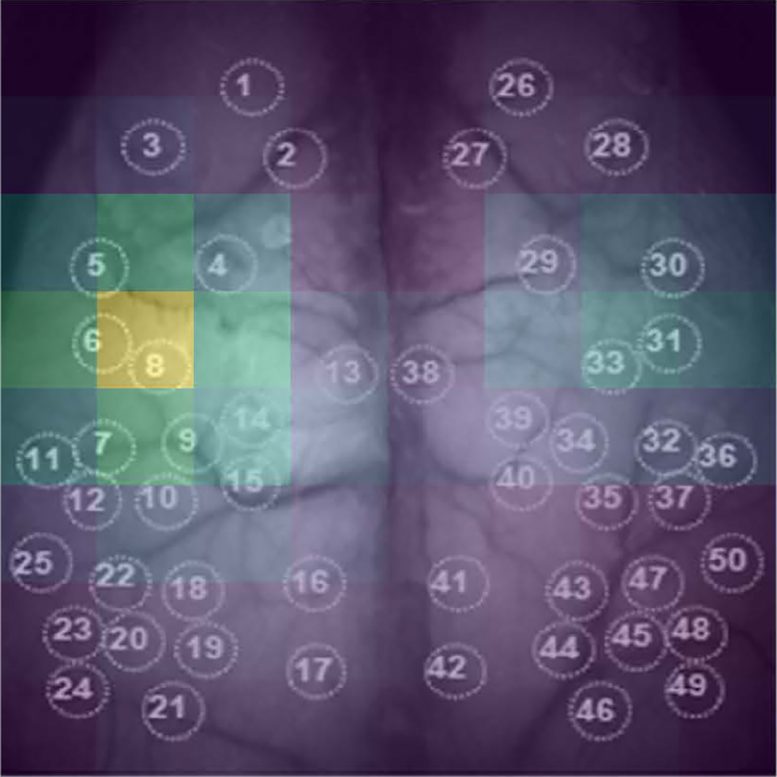
A new “end-to-end” deep learning method for the prediction of behavioral states uses whole-cortex functional imaging that do not require preprocessing or pre-specified features. Developed by medical student AJIOKA Takehiro and a team led by Kobe University’s TAKUMI Toru, their approach also allows them to identify which brain regions are most relevant for the algorithm (pictured). The ability to extract this information lays the foundation for future developments of brain-machine interfaces. Credit: Ajioka Takehiro
An AI image recognition algorithm can predict whether a mouse is moving or not based on brain functional imaging data. The researchers from Kobe University have also developed a method to identify which input data is relevant, shining light into the AI black box with the potential to contribute to brain-machine interface technology.
For the production of brain-machine interfaces, it is necessary to understand how brain signals and affected actions relate to each other. This is called “neural decoding,” and most research in this field is done on the brain cells’ electrical activity, which is measured by electrodes implanted into the brain. On the other hand, functional imaging technologies, such as fMRI or calcium imaging, can monitor the whole brain and can make active brain regions visible by proxy data.
Out of the two, calcium imaging is faster and offers better spatial resolution. But these data sources remain untapped for neural decoding efforts. One particular obstacle is the need to preprocess the data such as by removing noise or identifying a region of interest, making it difficult to devise a generalized procedure for neural decoding of many different kinds of behavior.
Breakthrough in Neural Decoding with AI
Kobe University medical student Ajioka Takehiro used the interdisciplinary expertise of the team led by neuroscientist Takumi Toru to tackle this issue. “Our experience with VR-based real-time imaging and motion tracking systems for mice and deep learning techniques allowed us to explore ‘end-to-end’ deep learning methods, which means that they don’t require preprocessing or pre-specified features, and thus assess cortex-wide information for neural decoding,” says Ajioka. They combined two different deep learning algorithms, one for spatial and one for temporal patterns, to whole-cortex film data from mice resting or running on a treadmill and trained their AI model to accurately predict from the cortex image data whether the mouse is moving or resting.
In the journal PLoS Computational Biology, the Kobe University researchers report that their model has an accuracy of 95% in predicting the true behavioral state of the animal without the need to remove noise or pre-define a region of interest. In addition, their model made these accurate predictions based on just 0.17 seconds of data, meaning that they could achieve near real-time speeds. Also, this worked across five different individuals, which shows that the model could filter out individual characteristics.
Identifying Key Data for Predictions
The neuroscientists then went on to identify which parts of the image data were mainly responsible for the prediction by deleting portions of the data and observing the performance of the model in that state. The worse the prediction became, the more important that data was. “This ability of our model to identify critical cortical regions for behavioral classification is particularly exciting, as it opens the lid of the ‘black box’ aspect of deep learning techniques,” explains Ajioka.
Taken together, the Kobe University team established a generalizable technique to identify behavioral states from whole-cortex functional imaging data and developed a technique to identify which portions of the data the predictions are based on. Ajioka explains why this is relevant. “This research establishes the foundation for further developing brain-machine interfaces capable of near real-time behavior decoding using non-invasive brain imaging.”
Reference: “End-to-end deep learning approach to mouse behavior classification from cortex-wide calcium imaging” by Takehiro Ajioka, Nobuhiro Nakai, Okito Yamashita and Toru Takumi, 13 March 2024, PLOS Computational Biology.
DOI: 10.1371/journal.pcbi.1011074
This research was funded by the Japan Society for the Promotion of Science (grants JP16H06316, JP23H04233, JP23KK0132, JP19K16886, JP23K14673 and JP23H04138), the Japan Agency for Medical Research and Development (grant JP21wm0425011), the Japan Science and Technology Agency (grants JPMJMS2299 and JPMJMS229B), the National Center of Neurology and Psychiatry (grant 30-9), and the Takeda Science Foundation. It was conducted in collaboration with researchers from the ATR Neural Information Analysis Laboratories.


